Export to Cytoscape
Instead of working with the Neo4j database directly, it is also possible to export a specific network to a graphml file or to a running Cytoscape instance with the commands below. Cytoscape is an open source platform for visualizing networks; if you have not yet installed it, download it from the Cytoscape homepage. Specify the target network with the -net flag. The new network is exported to the specified file path in -fp. The -cyto flag is used to tell mako to export the network to Cytoscape as well.
mako io -fp . -cf -net Genus_demo_1 -cyto -w
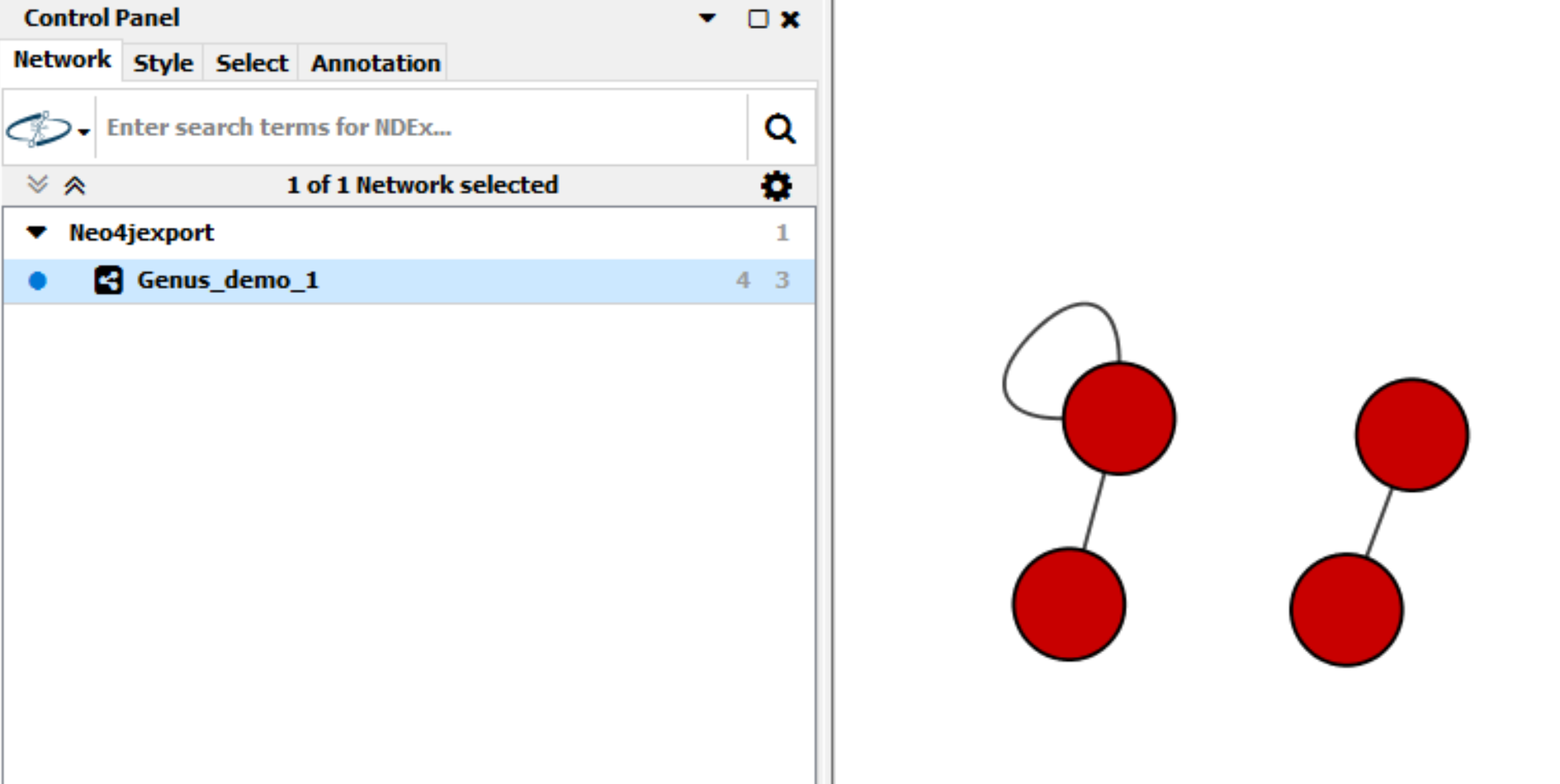
The Genus_demo_1 wnetwork as previously generated using the mako module; by specifying this network name, it is possible to export the network to Cytoscape. When doing so, mako converts the Neo4j representation of the network to a conventional network representation where edges are only relationships between nodes (Figure 6).