Calculate metadata correlations
The command below carries out hypergeometric tests to see if the metadata (skin vs gut) is linked to specific taxa. Note that this functionality is meant for exploratory analysis only and does not include multiple-testing corrections. Depending on the type of metadata available to you and the structure of your experimental design, more appropriate statistical methods may be available. The -var all parameter tells mako to run the tests across all variables.
mako metastats -fp . -cf -var all
If you access the Neo4j Browser (http://localhost:7475/browser/) and run the following query, you should be able to access all taxa that were linked to the skin or gut samples via the hypergeometric test:
MATCH p=(n:Taxon)--(:Property) RETURN p LIMIT 50
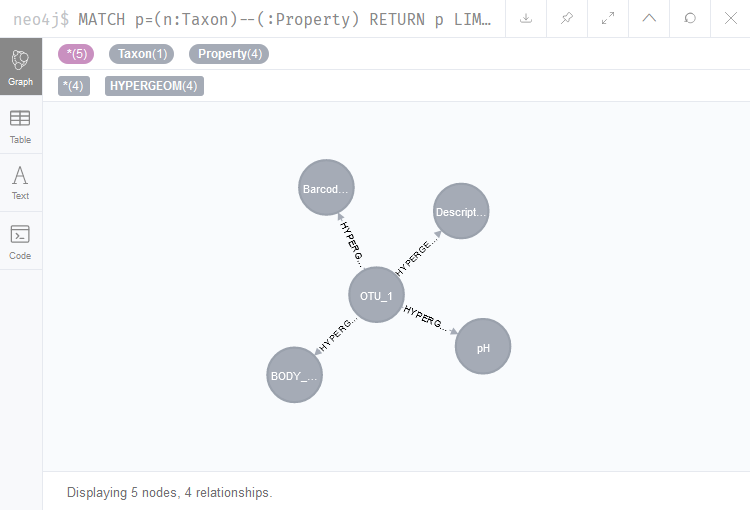
The query MATCH p=(n:Taxon)–(:Property) RETURN p LIMIT 50 returns 50 patterns consisting of Taxon nodes connected to Property nodes. In this case, the only possible connections matching this pattern were previously generated by the hypergeometric test, as the name of the relationships indicates (Figure 4). Here, the query result therefore shows that OTU_1 was the only taxon linked to a certain pH and body site, as it was connected to those nodes via a hypergeometric test.