Uploading networks
From a terminal, navigate to the location where you downloaded the network files. Now, only the network files and the file path need to be provided.
mako io -fp . -cf -net demo_1.graphml demo_2.graphml
If you access the Neo4j Browser (http://localhost:7475/browser/) and run the following query, you should be able to access all the nodes connected to edges:
MATCH p=(n:Edge)--() RETURN p LIMIT 50
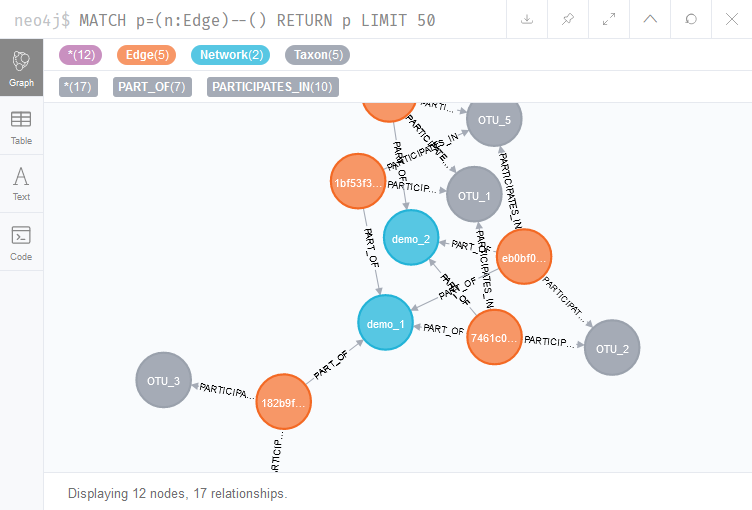
The query MATCH p=(n:Edge)–() RETURN p LIMIT 50 returns 50 patterns consisting of Edge nodes connected to any other node. Since Edge nodes can connect to Taxon nodes and to Network nodes, these are returned by the query (Figure 2).
In this case, the shown edges are part of the imported synthetic networks, the demo_1.graphml and the demo_2.graphml files. The interpretation of these networks therefore depends on the type of data that the grapmhl files were based on. If these were association networks, the orange Edge nodes would represent statistically significant links between different OTUs (and therefore do not represent observations of interactions). However, if they were metabolic interaction networks, each Edge node could represent an exchange of metabolites.