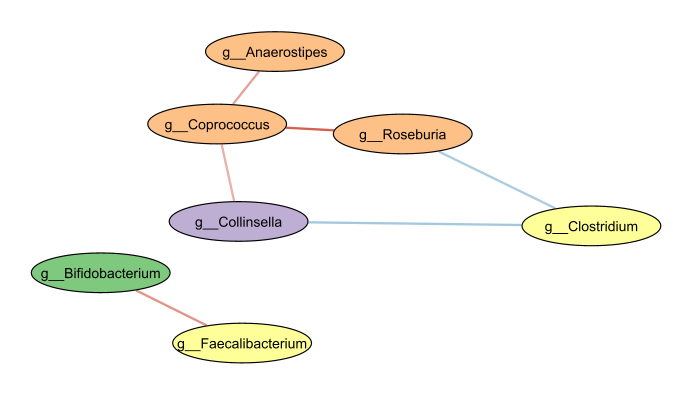
Associations linked to literature-validated interactions
With both networks available via the Neo4j database, we can start investigating associations that could be related to molecules of interest, like butyrate. Are there microbes co-occurring that could produce butyrate from starch?
Since this is not a metabolic network, we will need to consider which intermediates are required for the production of butyrate. Specifically, we will look at cross-feeding interactions that are mediated via the secretion of acetate and lactate. We need to look at all species that are able to degrade starches and produce acetate and lactate, and see if there are any associations between these and species that produce butyrate from consuming acetate and lactate.
Before we begin, we need to collect all names of the metabolites of interest. For example, starch has quite a long name: “Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)”. Butyrate and acetate are simply listed as “Butyrate” and “Acetate”, while lactate is in the NJS16 network under the name “L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)”.
First, we will write a query that finds species consuming starch, and producing (or consuming and producing) acetate. The first node in the query finds the starch metabolite. The relationship specifies that the microbe should be able to degrade starch. The second relationship is assigned a variable, r, so we can use a WHERE clause to state that the microbe should be able to produce acetate (but may also consume it). Note that you can copy the query and run it from Neo4j Browser, but you can also choose to run it via the driver.query() function.
Note that we included the LIMIT 50 clause to make sure only limited results are returned; if you try to return all results, the Neo4j Browser may crash.
MATCH p=(:Metabolite {name: 'Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)'})
-[:QUALITY_OF {interaction: 'macromolecule_degradation'}]
-(:Microbe)-[r]-(:Metabolite {name: 'Acetate'})
WHERE r.interaction IN ['production', 'consumption_production']
RETURN p LIMIT 50
We can easily adapt this query to find producers of lactate as well. To do so, we need to do the same thing we did with the relationship; assign a variable to the second metabolite and state in the WHERE clause that it needs to be either acetate or lactate.
MATCH p=(:Metabolite {name: 'Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)'})
-[:QUALITY_OF {interaction: 'macromolecule_degradation'}]
-(:Microbe)-[r]-(b:Metabolite)
WHERE r.interaction IN ['production', 'consumption_production']
AND b.name in ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate']
RETURN p LIMIT 50
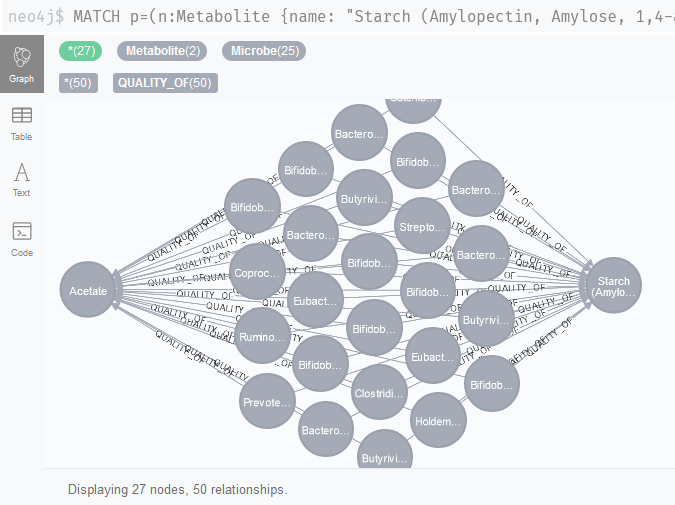
Indeed, if we run these queries in the browser, some known suspects like Bifidobacterium, a well-studied cross-feeding bacterium, show up (Figure 3).
We next need to identify microbes that can produce butyrate from either acetate or lactate. We can do this simply by changing the relationship values and replacing the starch node.
MATCH p=(:Metabolite {name: 'Butyrate'})
-[q]-(:Microbe)-[r]-(b:Metabolite)
WHERE q.interaction IN ['production', 'consumption_production']
AND r.interaction IN ['consumption', 'consumption_production']
AND b.name in ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate']
RETURN p LIMIT 50
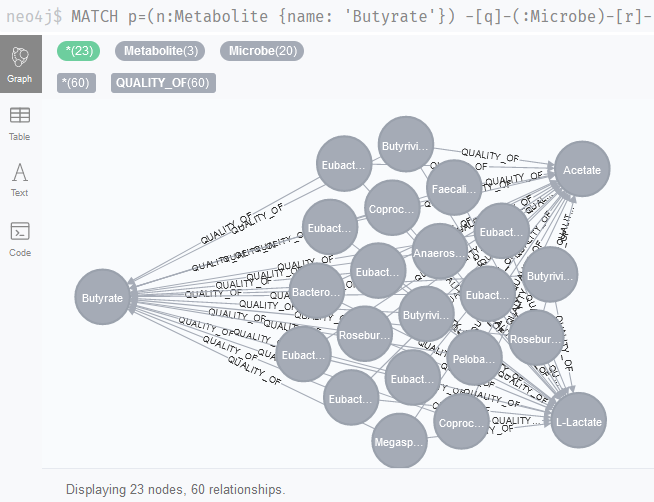
And again, we get some known offenders; Roseburia, Faecalibacterium and Eubacterium are all known to be responsible for butyrate production in the human gut (Figure 4).
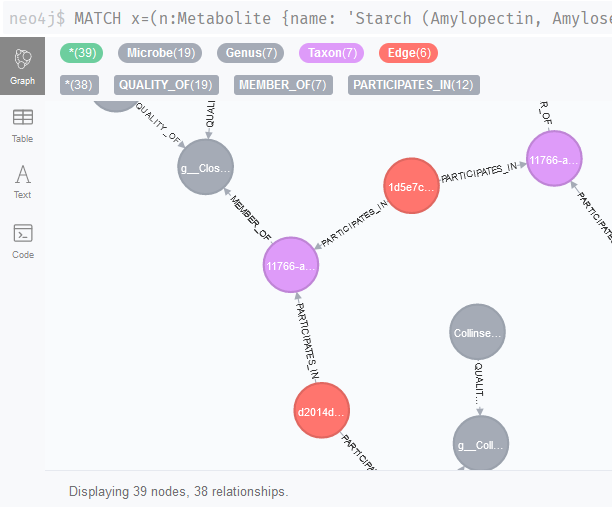
The next thing we need to do, is to actually combine these queries and find all associations that could be linked to these genera. We can do this by including multiple match queries.
MATCH x=(n:Metabolite {name: 'Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)'})
-[:QUALITY_OF {interaction: 'macromolecule_degradation'}]
-(:Microbe)-[r]-(b:Metabolite)
WHERE r.interaction IN ['production', 'consumption_production']
AND b.name IN ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate']
MATCH y=(:Metabolite {name: 'Butyrate'})-[c]-(:Microbe)-[d]-(e:Metabolite)
WHERE c.interaction IN ['production', 'consumption_production']
AND e.name in ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate']
RETURN x, y LIMIT 100
Of course, this only gives us all the Microbe nodes and we need the associations. Therefore, we can use a WITH clause to continue the query and find all associations. We use the i and j parameters to refer to the Microbe nodes and then continue the query; since we connected microbes to genera and genera are connected to taxa, we can find all associations linked to these literature-validated interactions with a relatively straightforward pattern.
MATCH x=(n:Metabolite {name: 'Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)'})
-[:QUALITY_OF {interaction: 'macromolecule_degradation'}]
-(i:Microbe)-[r]-(b:Metabolite)
WHERE r.interaction IN ['production', 'consumption_production']
AND b.name IN ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate']
MATCH y=(:Metabolite {name: 'Butyrate'})
-[c]-(j:Microbe)-[d]-(e:Metabolite)
WHERE c.interaction IN ['production', 'consumption_production']
AND d.interaction IN ['consumption', 'consumption_production']
AND e.name in ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)','Acetate']
WITH i, j MATCH p=(i)--(:Genus)--(:Taxon)--(:Edge)--(:Taxon)--(:Genus)--(j)
RETURN p LIMIT 100
We only need to add one last trick before we can easily export this network to Cytoscape: we are going to connect these associations to a Network node, so we can export this network to Cytoscape. For this, we need to use the driver, because the Neo4j Browser may not be able to run such a large query.
Before you run the driver.export_cyto() command, make sure to have Cytoscape open!
driver.write("MERGE (n:Network {name: 'Butyrate_network'}) RETURN n")
butyrate_query = "MATCH x=(n:Metabolite {name: 'Starch (Amylopectin, Amylose, 1,4-alpha-D-Glucan, Pullulan, Resistant starch)'})-" \
"[:QUALITY_OF {interaction: 'macromolecule_degradation'}]-(i:Microbe)-[r]-(b:Metabolite) " \
"WHERE r.interaction IN ['production', 'consumption_production'] " \
"AND b.name IN ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate'] " \
"MATCH y=(:Metabolite {name: 'Butyrate'})-[c]-(j:Microbe)-[d]-(e:Metabolite) " \
"WHERE c.interaction IN ['production', 'consumption_production'] " \
"AND d.interaction IN ['consumption', 'consumption_production'] " \
"AND e.name in ['L-Lactate ([S]-Lactate, Lactate, D-Lactate, [R]-Lactate)', 'Acetate'] " \
"WITH i, j MATCH p=(i)--(:Genus)--(:Taxon)--(z:Edge)--(:Taxon)--(:Genus)--(j) RETURN z"
results = driver.query(butyrate_query)
edge_names = [{"name": x['z']['name']} for x in results]
query = "WITH $batch as batch " \
"UNWIND batch as record " \
"MATCH (a:Edge {name:record.name}), (b:Network {name: 'Butyrate_network'})" \
"MERGE (a)-[r:PART_OF]-(b) RETURN r"
driver.write(query, batch=edge_names)
driver.export_cyto(networks=['Butyrate_network'])
After we format the network, we can see that there is indeed a small number of associations between taxa that are able to degrade starch and produce the substrates required for other taxa to produce butyrate. Although we do not have evidence that these taxa are actually producing butyrate in concert, the positive associations between Coprococcus, Anaerostipes and Roseburia, as well as the associations between Bifidobacterium and Faecalibacterium do match the literature, suggesting that these taxa could indeed be producing butyrate in this particular collection of microbiome samples.