Network motifs
Network motifs are a type of recurring pattern or subgraph that can be found in larger networks. In transcription networks, some motifs lead to dynamics that are crucial for organism functionality (Alon, 2007). However, the motifs described by Alon are directed: they show whether gene A affects gene B, or the other way around. Consequently, not all insights from Alon’s and related work may translate to microbial association networks. Other methodological issues are also at play: edges in transcriptional networks are much more likely to reflect actual interactions compared to edges in association networks, since many are inferred from experimental work or can be verified that way.
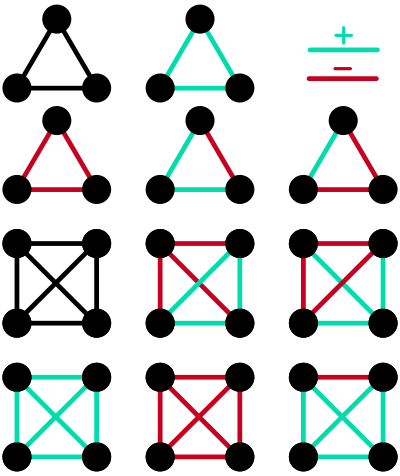
Consequently, our aim with this case study was to show the distribution of certain network motifs in a large data set, to generate initial hypotheses on the sorts of motifs that we might find. Specifically, we looked for densely connected 3- and 4-node cliques. Cliques are motifs where all nodes in the clique are connected to all other cliques. We were interested in seeing if specific combinations of edge weights in cliques were more common than other combinations, and whether this was different across EMPO_2 annotations (Animal, Non-saline, Plant, Saline).
In this case study, we will first explain some functions that we need to process Cypher query results. Next, we will go through the queries that we used to access the database. Finally, we will explain how we visualized these results in R. We assume that a running Neo4j database with all 60 data sets is already available (see the Qiita case study for more information).